Sources of Information:
Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter
Jozef Mravec1,2, Petr Sku°pa3, Aure´lien Bailly4, Kla´ra Hoyerova´3, Pavel Krˇecˇek3, Agnieszka Bielach1, Jan Petra´sˇek3,5, Jing Zhang1,2, Vassilena Gaykova2, York-Dieter Stierhof2, Petre I. Dobrev3, Katerˇina Schwarzerova´5, Jakub Rolcˇı´k6, Daniela Seifertova´3, Christian Luschnig7, Eva Benkova´1, Eva Zazˇı´malova´3, Markus Geisler4 & Jirˇı´ Friml1,8
The article focuses on finding PIN 5 role and its location in cells.
PICTURE 1 :
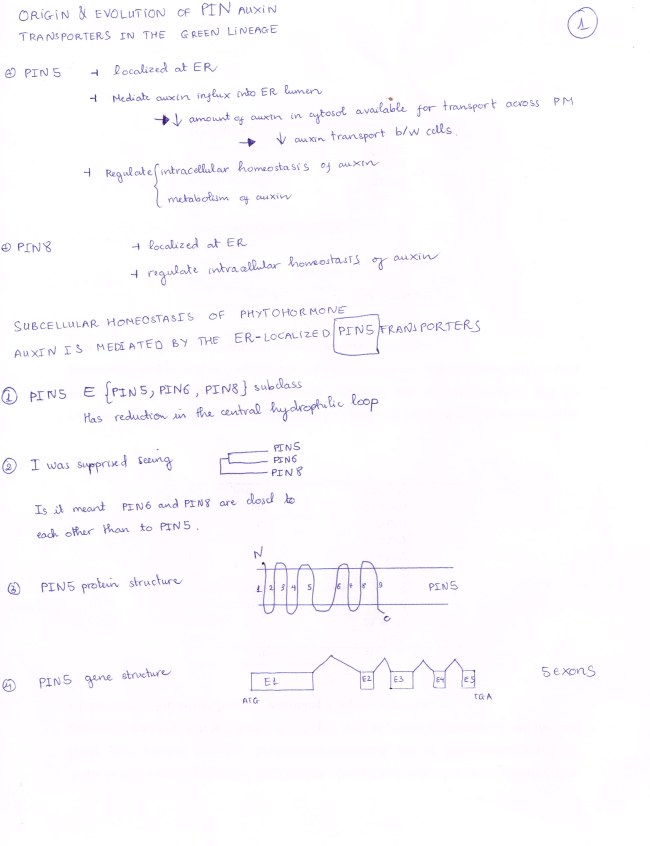
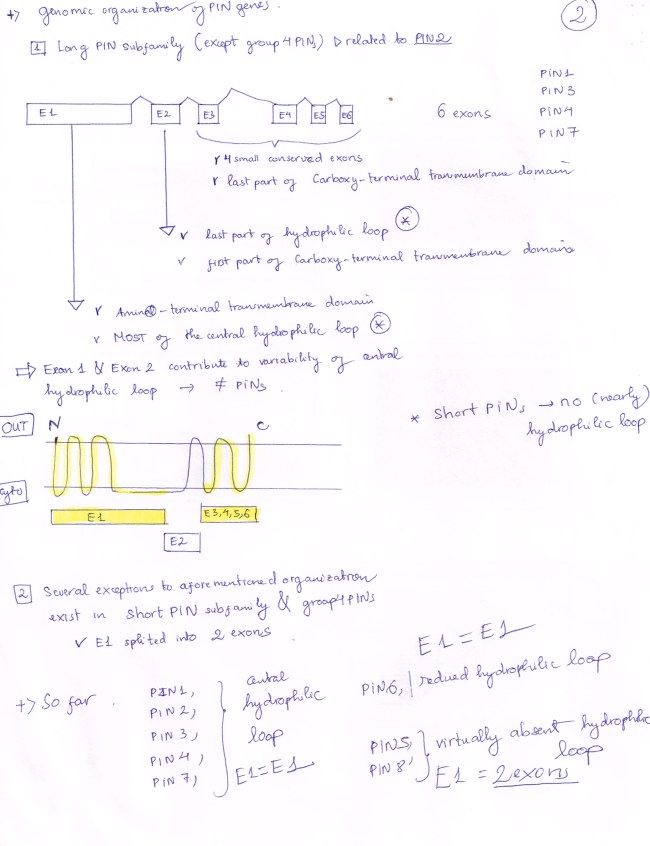
PIN5 gene is quite different from the consensus that SELF-REVIEW 1 stated: PIN 5 has only 5 Exons, with ATG and TGA as its start codon and stop codon respectively.
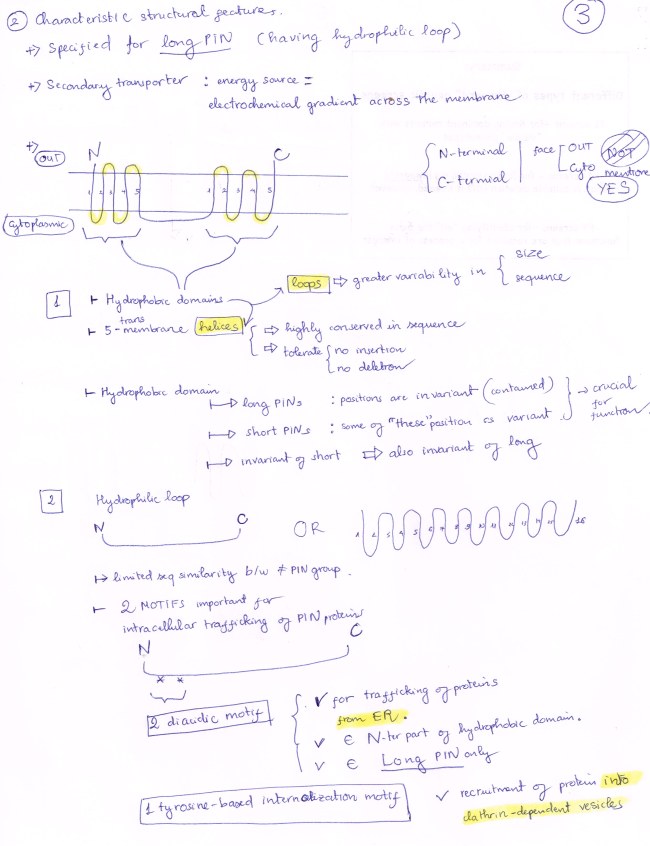
PIN 5 protein has a FIVE-transmembrane hydrophonic domain,
a FOUR-transmembrane hydrophobic domain, and
a VERY SHORT hydrophilic loop (virtually absent)
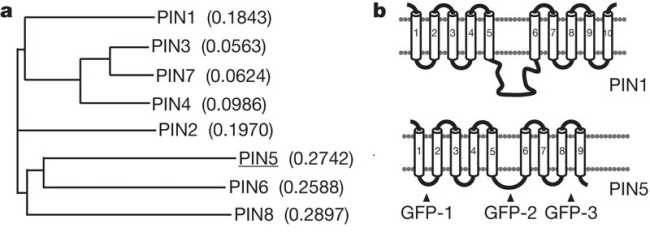
PICTURE next : is actually taken directly from the aforementioned article. The big surprise comes from it is the phylogenetic branchs of PIN 5, PIN 6, and PIN 8. It seems that PIN 6 and PIN 5 are more closely related to each other than to PIN 8. Anyway, here is the picture:
PICTURE 2
In Auxin Transport Assay in Heterologous Yeast System, it is interesting seeing PIN5-HA in the Plasma Membrane (lately it is interpreted as result of PIN 5 ectopic expression). as expected from a member of PIN family, PIN 5 can transport auxin through PM.
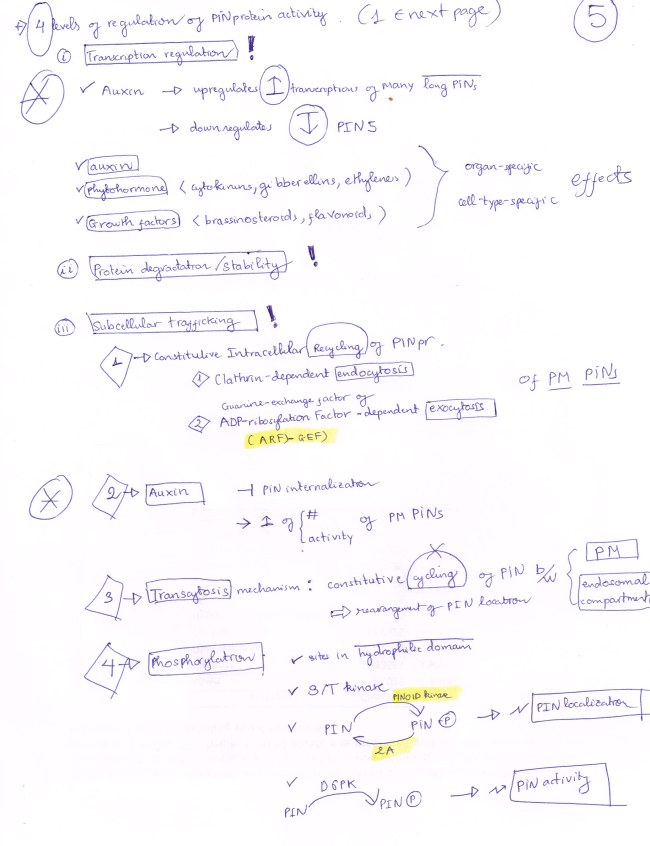
PIN 5 is auxin-regulated broadly expressed auxin transporter.
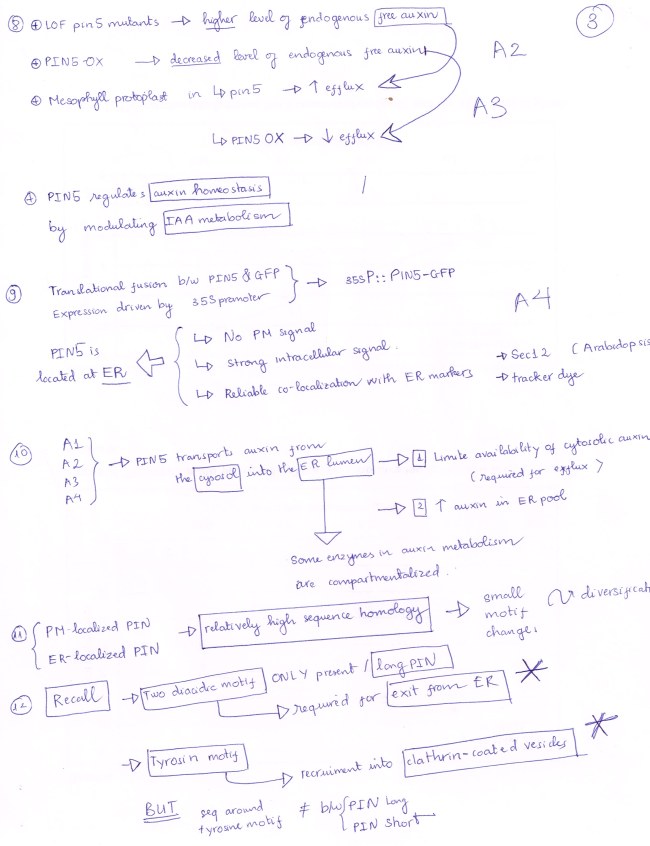
T-DNA insertion generates Loss-of-Function pin5 mutants.
Gain-of-Function mutant is generated by employing STRONG PROMOTER to drive OVERexpression of PIN5.
PICTURE 3
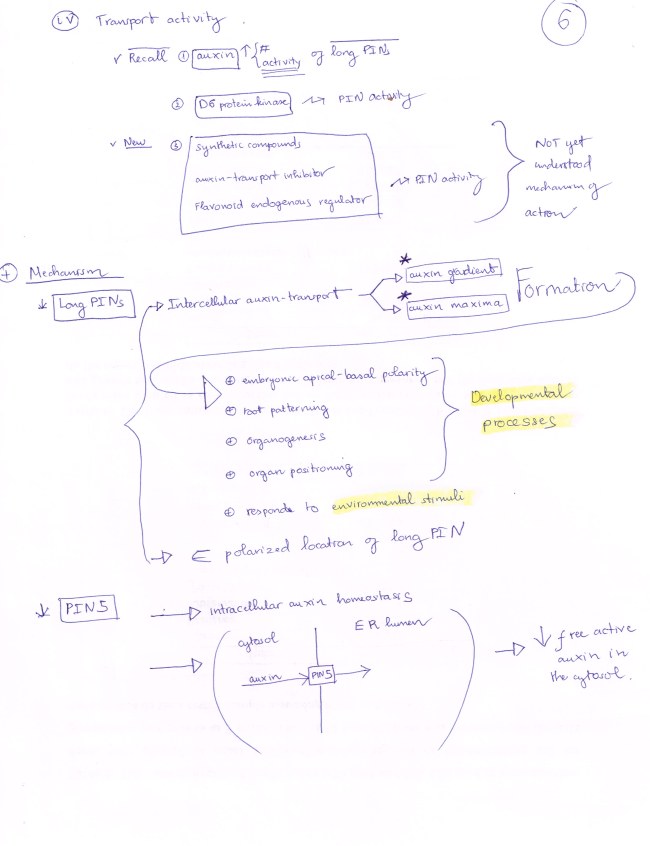
NEW but OLD: PIN 5 protein is located at ER membrane (also PIN 6, PIN 8 in Arabidopsis).
PIN 5 transport auxin from CYTOSOL into ER LUMEN, hence limiting the availability of cytosolic free auxin for auxin export, and increasing level of auxin in ER pool for auxin metabolism.
The relatively high sequence homology between PM-localized PIN and ER-localized PIN suggests the diversification of PIN is due to small motif changes.
By the way, because we mention MOTIF, I will talk about two types of motifs found in PIN protein:
(ONE) : Diacidic Motif: Two of them are found in LONG PIN only, and they are signal for PIN exit from ER.
(TWO): Tyrosine Motif: Found in all members of PIN family, recruits PIN into Clathrin-Coated Vesicles. HERE, the special thing does not come from the motif itself, in stead it comes from the varied sequences around the site of this motif.
And last, the EXTRA picture (fun scan)
OK! I can call it the end of today 🙂